当前位置:网站首页>2022 | Sample Efficiency Matters: A Benchmark for Practical Molecular Optimization
2022 | Sample Efficiency Matters: A Benchmark for Practical Molecular Optimization
2022-07-21 07:38:00 【Stunned flounder (】
2022 | Sample Efficiency Matters: A Benchmark for Practical Molecular Optimization

Paper: https://arxiv.org/abs/2206.12411
Code: https://github.com/wenhao-gao/mol_opt
PMO: Compare 25 Sample efficiency of molecular optimization methods
Molecular optimization is a basic goal of Chemical Science , It is the core interest of drug and material design . In recent years , Significant progress has been made in solving challenging problems in all aspects of computational molecular optimization , High effectiveness is emphasized 、 diversity , And recent composability . Despite these advances , Many papers report the results of trivial or self-designed tasks , This poses additional challenges to directly evaluate the performance of the new method . Besides , Optimization efficiency of samples (Oracle Number of molecules evaluated ) Rarely discussed , Although this is a necessary consideration for practical discovery applications .
To fill this gap , The author proposes an open source baseline for molecular optimization (PMO), To facilitate the replication evaluation of molecular optimization algorithms . And in-depth study 25 A molecular design algorithm stay 23 Species task Performance on . Experiments show that , In the Limited oracle Under the circumstances , Most of the most advanced methods cannot surpass previous algorithms , And the existing algorithms can not effectively solve some molecular optimization problems . therefore , From the choice of Optimization Algorithm 、 Molecular assembly strategies and oracle To share the impact on optimization performance , Provide information for future algorithm development and baseline testing . All the codes can be found here :https://github.com/wenhao-gao/mol_opt
Introduce
Although exciting progress has been made in this field and new methods have been proposed , But how these algorithms compare with each other is still unclear . There are at least three problems :
- Lack of right oracle consider : Many papers are not reported oracle How many times has it been called to achieve the reported results .
- Some papers only report trivial oracles result , Such as quantitative estimation of drug similarity (QED) Or punish octanol - Water partition coefficient (LogP); Other papers even introduced new self-designed tasks , This confused the comparison with previous work .
- Randomness : Many algorithms are not deterministic , And show significant operational differences , Therefore, it is necessary to report the results of several independent tests .
therefore , The author proposes a new repeatable large-scale experimental model (PMO). Yes 23 Kind of oracle Conduct 25 Method benchmark , Each method is tuned , And conducted many independent tests . Consider the combination of optimization ability and sample efficiency , Limit orcale Call the number (10000), And use AUC To measure model performance .PMO Benchmarks will make molecular optimization easier to obtain and reproducible , So as to promote the progress of the algorithm , Finally, molecular optimization technology will be more widely used in the workflow of experimental drugs and material discovery .
Algorithm
Molecular optimization methods have two parts :
- Molecular assembly strategy : Molecular assembly implicitly defines the chemical space explored by the algorithm , The optimization algorithm determines how to explore in this space . The assembly of molecules is similar to the expression of molecules in machine learning , There are mainly the following :(1)SMILES character string ;(2)SELFIES character string ;(3) Based on atoms (atom) The molecular diagram of (molecular graph);(4) Based on groups (fragment) The molecular diagram of ( May include atoms );(5) Synthesis diagram based on chemical reaction .
- An optimization method : It mainly includes the following 9 class , genetic algorithm(GA, Genetic algorithm (ga) ), Monte Carlo Tree Search(MCTS, Monte carlo tree search ), Bayesian optimization (BO, Bayesian optimization ), variational autoencoder(VAE, Variational self encoder ), generative adversarial network(GAN, Generative antagonistic network ),score-based model (SBM, Probability model based on score ), hill climbing (HC, Climbing algorithm ), reinforcement learning(RL, To enhance learning ), gradient ascent(GRAD, The gradient rises ).
The following table summarizes the current molecular design methods based on the classification of assembly strategies and optimization methods , Including but not limited to the methods included in the baseline .
The experimental conclusion
Oracle: In order to test the generality of the method , Our goal is to cover a wide range of pharmaceutical related Oracle function .
Data sets : Use ZINC 250K Data sets , The dataset contains data from ZINC About... Sampled in the database 250K molecular
Evaluation indicators : In order to consider both optimization capability and sample efficiency , We report top-K The area under the curve of the average attribute value (AUC) And oracle Call the number (AUC top-K) Comparison of , As the main index to measure performance .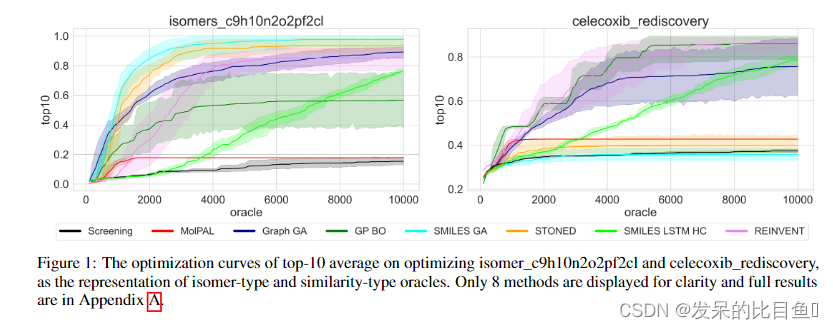
The efficiency of the sample
All current methods , Except in some very simple oracle outside , Cannot be called hundreds of times oracle To complete the optimization . This shows that all current molecular optimization algorithms are difficult to directly take experimental measurements on small molecules oracle Optimize molecules . By comparison top-X and AUC Top-X Ranking , You can see some in Top-X Which method of performance is acceptable in AUC Top-X I didn't do well in the game , Explain the previously considered strong Algorithm , The efficiency of using samples is not strong , Proved AUC The significance of this indicator .
Traditional methods are still powerful
The top two comprehensive ranking methods are REINVENT( be based on SMILES Reinforcement learning methods ),Graph GA( Genetic algorithm based on molecular graph ). Neither method has been published in AI Summit meeting , In contrast, many recent years AI The paper of the top meeting is not well realized .
The following table is based on the average AUC Top-10 Of 10 The performance of the best molecular optimization method . The experiment report 5 An independent test AUC Top-10 Mean and standard deviation . The best model in each task is marked in bold .



smile There are no obvious shortcomings
It is found in the comparative test ,SMILES In most cases, it is better than SELFIES. The author believes that the main reason is
- The current language model can be learned SMILES The grammar of , Ensure that the generated molecules have enough effective molecules , So it's smoothed SELFIES The advantages of .
- Through the case study, it is found that , In fact, SELFIES There are also grammatical errors in , It's just SMILES If there is a syntax error in, an error will be reported , And in the SELFIES Cover up your mistakes , Although the resulting molecules are effective in bond valence , But it did not bring effective chemical space exploration .
A model-based approach may be more effective , But it needs careful design
model-based The optimization method is to train a predictive model As truth oracle substitute , To optimize , To achieve the purpose of more efficient optimization . On the other hand ,model-based The optimization method of needs more careful design , Simply add one GNN Training does not necessarily have the desired improvement .

Different types of methods are more suitable for different tasks
The authors found that, for example, in some isomer based tasks , Genetic algorithm method based on string ( such as SMILES-GA,STONED be based on SELFIES) The effect is better. . In the task based on similarity , The effect of these methods is not ideal . This shows that different optimization methods have different adaptability in different types of tasks , Depending on oracle Of landscape.
When reporting results, you need to re optimize the parameters and run multiple times
By adjusting the super parameters, we get a comparison with REINVENT Original paper Better results in , It shows that when comparing different models , The importance of parameter adjustment .
Reference resources
https://mp.weixin.qq.com/s/mgO16tuLVaovLyJA64agrw
边栏推荐
猜你喜欢
随机推荐
G024-DB-GS-INS-01 OpenEuler 部署 OpenGauss(单实例)
MapStruct - Consider defining a bean of type in your configuration.
2022 | Sample Efficiency Matters: A Benchmark for Practical Molecular Optimization
Kusionstack open source | exploration and practice of kusion model library and tool chain
OSPF comprehensive experiment
年度重磅!华为云2021应用构建技术实践精选集,七大领域400页+云上开发宝典 | 云享·书库 No.02 期推荐(附免费下载)
上海文旅局局长:安全是文旅业的生命线,正抢抓元宇宙新赛道
R语言epiDisplay包的idr.display函数获取泊松回归poisson模型的汇总统计信息(初始事件密度比IDR值、调整事件密度比IDR值及其置信区间、Wald检验的p值和似然比检验的p值
放弃免费Inoreader 自建RSS阅读器—Tiny Tiny RSS和FreshRSS
高能同步辐射光源科学数据管理策略研究与应用
C and pointer Chapter 1 lexical "trap" 1.3 "greedy method" in lexical analysis
Thymeleaf uploads files in web pages
动手搭建一个三个节点的eureka集群
越级产品再升级,2023款吉利星瑞11.37万元起售
LeetCode#437 路径总和3
UIScrollView 和 UIPageControl 实现启动滑动图[通俗易懂]
MapStruct - Consider defining a bean of type in your configuration.
HCIP笔记第十天
推荐一个 WordPress 付费主题站
自动推理的逻辑05--谓词演算









