当前位置:网站首页>RNA 23. Risk factor association diagram of Cox model of expressed genes in SCI articles (ggrisk)
RNA 23. Risk factor association diagram of Cox model of expressed genes in SCI articles (ggrisk)
2022-07-21 04:46:00 【Huanfeng gene】
Click to follow , Huanfeng gene
Huanfeng gene
Bioinformatics analysis ,SCI Writing articles and learning basic knowledge of biological information :R Language learning ,perl Basic programming ,linux System commands ,Python Meet a better you
130 Original content
official account
The tutorial of Huanfeng gene not only teaches you how to use , We will also analyze some related articles regularly , Learning tutorials is just the foundation , But if the analysis results are integrated into the article, it is the purpose , I think these tutorials are not bad , And you analyzed the good results according to our tutorial and sent an article. Remember to inform us , And thank us in the article !
The English name of the company :Kyoho Gene Technology (Beijing) Co.,Ltd.
Huanfeng gene official account launched a course of transcriptome analysis and clinical prediction model , Teachers who need students' letters can contact us !
First, take a look at the transcription analysis tutorial, which is summarized as follows :
RNA 1. Gene expression things – be based on GEO
RNA 2. SCI The article is based on GEO Of differentially expressed genes limma
RNA 3. SCI The article is based on T CGA Differentially expressed genes DESeq2
RNA 4. SCI The article is based on TCGA Differential expression edgeR
RNA 5. SCI Differential gene expression in the article MA chart
RNA 6. Differential gene expression -- Volcanic map (volcano)
RNA 7. SCI Gene expression in the article —— Principal component analysis (PCA)
RNA 8. SCI Differential gene expression in the article – Thermogram (heatmap)
RNA 9. SCI Gene expression in the article GO notes
RNA 10. SCI Gene expression is enriched in the article –KEGG
RNA 11. SCI Gene expression is enriched in the article GSEA
RNA 12. SCI The calculation method of tumor immune infiltration in this article CIBERSORT
RNA 13. SCI Of differentially expressed genes in the article WGCNA
RNA 14. SCI Of differentially expressed genes in the article Protein protein interaction network (PPI)
RNA 15. SCI One of the fusion genes in the article FusionGDB2
RNA 16. SCI The visualization of fusion genes in this article
RNA 17. SCI Screening in the article Hub gene (Hub genes)
RNA 18. SCI Analysis of gene set variation in the article GSVA
RNA 19. SCI Unsupervised clustering method in the article (ConsensusClusterPlus)
RNA 20. SCI Single sample immune infiltration analysis in the article (ssGSEA)
RNA 21. SCI Single gene enrichment analysis in the article
RNA 22. SCI In this paper, the stromal cells and immune cells of malignant tumor tissues are estimated based on expression (ESTIMATE) original The modified
RNA 23. SCI The risk factor association diagram of the expressed gene model in the article (ggrisk)
The clinical prediction model is summarized as follows :
Topic 1. _ clinical _ Conventional thinking of biomarker Shengxin analysis
Topic 2. Survival analysis Kaplan-Meier
Topic 3. SCI The first table of the article – Baseline table
Topic 4. _ clinical _ Prediction model construction Logistic Return to
Topic 5. Sample size determination and segmentation
Topic 6 Counting variable Poisson regression
Topic 7. _ clinical _ prediction model –Cox Return to
Topic 8. _ clinical _ prediction model -Lasso Return to
Topic 9. SCI The second table of the article — Single factor regression analysis table
Topic 10. Single factor Logistic regression analysis — Univariate analysis table
Topic 11. SCI Multivariate screening — single / Multifactor table
Topic 12 _ clinical _ prediction model — Nomograph (Nomogram)
Topic 13. _ clinical _ prediction model — Consistency index (C-index)
Topic 14. _ clinical _ Calibration curve of prediction model (Calibration curve)
Topic 15. _ clinical _ Decision curve of prediction model (DCA)
Topic 16. _ clinical _ Receiver operation characteristic curve of prediction model (ROC)
Topic 17. Missing value recognition and visualization of clinical prediction models
Topic 18. Missing value interpolation method of clinical prediction model
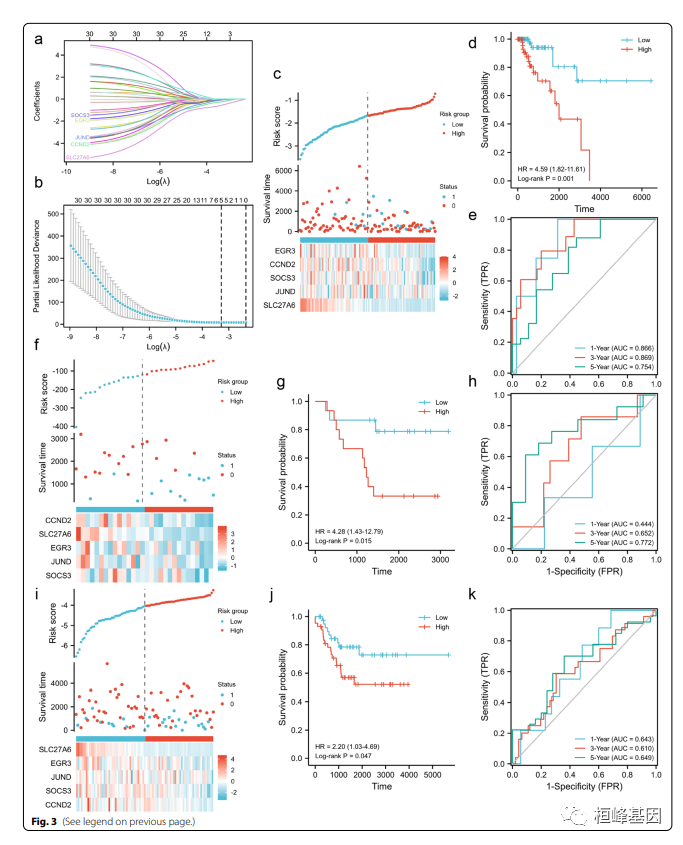
Yesterday I shared an article IF:4+ Iron metabolism and immune related gene markers predict clinical outcomes and molecular characteristics of triple negative breast cancer , A teacher asked questions , Say... In the article Fig.3 c,f,i How to draw the correlation diagram of risk factors ? In fact, there are ready-made software packages , Super easy , Let's see !!!
Preface
Risk map is probably one of the most commonly used graphs for tumor gene data analysis . We can draw the following two conclusions : Compare the prediction results of the model with the actual survival situation to observe whether the survival rate of the high-risk group is lower than that of the low-risk group , Whether the survival time is shorter than that of the low-risk group . The other is to compare the heat map and the scatter diagram , Look at the correlation between predictors and results . Risk factor correlation diagrams are common in cox Risk proportion model , It is commonly seen in the following figure, which shows the combination of three figures , It is shown by cox The proportion of high-risk and low-risk groups distinguished by the risk model 、 Time to live ( raw / die ) And gene expression of concern ( It is usually the genes involved in model construction ) Differences in distribution . Of course, when the model does not involve gene expression , Often, the risk factor correlation diagram only shows the upper two parts .
Software installation
In the absence of this ggrisk Before the bag , Drawing still requires that each diagram be drawn separately in the group diagram , There is now a ggrisk The software package is very convenient , Read in the data and you can get the result , and ABC The picture numbers are all there , Open not happy ?
if (!require(ggrisk)) install.packages("ggrisk")
library(ggrisk)
library(survival)
library(survminer)
data fetch
We use ggrisk Package built-in data set LIRI Liver cancer from Japan ICGC database , Including time 、 Events and four genes were tested .
ICGC Liver Data from Japan Description This data is a liver cancer data from Japan Data released in ICGC database (Link). It cantains time, event and four genes. An object of class data.frame with 232 rows and 6 columns.
data(LIRI)
head(LIRI)
## time status ANLN CENPA GPR182 BCO2
## 1 3.0410959 1 6.821354 3.0366550 0.00000000 0.2248344
## 2 2.5479452 0 1.073527 0.4654169 0.17895040 5.8924860
## 3 4.0273973 0 2.579530 0.7732644 0.06809686 3.5994330
## 4 0.1643836 1 14.183630 7.7239000 0.03749626 1.1194870
## 5 0.8219178 0 3.588320 2.3237710 0.16762610 2.6660850
## 6 2.8767123 0 6.079665 3.6674980 0.21788230 0.7691067
# Construct multifactor using four genes cox The regression model :
Example operation
1. structure COX Return to
We use four genes to construct multifactor cox The regression model :
library(rms)
fit <- cph(Surv(time, status) ~ ANLN + CENPA + GPR182 + BCO2, LIRI)
2. Risk factor linkage map drawing
ggrisk(fit, cutoff.value = "median", cutoff.x = 145, cutoff.y = -0.8)

3. Adjust the risk score cutoff And position
ggrisk(fit,
cutoff.value='cutoff', # Optional ‘median’, ’roc’ or ’cutoff’
cutoff.x = 150, #“cutoff” The horizontal position of the text
cutoff.y = -1#“cutoff” The vertical position of the text
)
## --
## ==
## Combination: 185
## Combination: 43

4. Other parameter settings
Other parameter settings include color , label , Modification of drawing elements such as legend , Specific instructions can be used directly ?ggrisk Check usage .
ggrisk(fit,
code.highrisk = 'High Risk',# High risk label , The default is ’High’
code.lowrisk = 'Low Risk', # Low risk label , The default is ’Low’
title.A.ylab='Risk Score', #A chart y Axis name
title.B.ylab='Survival Time(year)', #B chart y Axis name , Pay attention to distinguish between year month day
title.A.legend='Risk Group', #A Figure legend name
title.B.legend='Status', #B Figure legend name
title.C.legend='Expression', #C Figure legend name
relative_heights=c(0.1,0.1,0.01,0.15), #A、B、 Heat map annotation and heat map C The relative height of
color.A=c(low='green',high='red'),#A The color of the midpoint in the figure
color.B=c(code.0='green',code.1='red'), #B The color of the midpoint in the figure
color.C=c(low='green',median='white',high='red'), #C The color of the heat map
vjust.A.ylab=1, #A In the figure y Axis label to y Distance of coordinate axis , The default is 1
vjust.B.ylab=2#B In the figure y Axis label to y Distance of coordinate axis , The default is 2
)

5. Assign heat maps to show genes
We can also specify which genes to display the heat map , as follows :
ggrisk(fit, heatmap.genes = c("GPR182", "CENPA", "BCO2"))

6. Don't show the heat map
Show only the scatter chart , Don't show the heat map , Use it directly two_scatter() function , as follows :
two_scatter(fit, cutoff.value = "median", cutoff.x = 142, cutoff.y = -0.5)

Is this very simple , Follow the Huanfeng gene tutorial , Cast a successful you , Remember to pay attention to us , If you find it difficult to make a letter , Come and contact Huanfeng gene !!!
References:
- Li XF, Fu WF, Zhang J, Song CG. An iron metabolism and immune related gene signature for the prediction of clinical outcome and molecular characteristics of triple-negative breast cancer. BMC Cancer. 2022;22(1):619. Published 2022 Jun 7. doi:10.1186/s12885-022-09679-x
边栏推荐
- What is the easiest explanation for SaaS? Just read this one
- DNA 11. 识别肿瘤蛋白质三维结构上突变热点(HotSpot3D)
- RNA 24. SCI文章中基于TCGA的免疫浸润细胞分析的在线小工具——TIMER
- C语言基本概念——每天一遍小知识
- Operation skills of markdown in jupyter
- 动态调试JS代码
- Game improvement of smart people: Lesson 2 of Chapter 3: number of factors (dcount)
- Installation and management procedures
- Solve tensoflow2 No module named:tensorflow contrib
- C語言基本概念——每天一遍小知識
猜你喜欢

Progress report of Zhongguancun Software Park Project

Dynamic routing protocol rip experiment

Become a blogger in a year? To! Struggling oneself

DNA 9. 揭秘肿瘤异质性与TMB, MSI之间的相关性

Principle and protection of DOM XSS

DNA 7. Genome copy number variation analysis and visualization (gistic2.0)

超全干货:数据可视化的设计总结,工具,技术要点统统都有

学会如何选择图表类型,小白也能玩转数据分析

Account and authority management

Three handshakes and four disconnects in TCP
随机推荐
Keep updating the article I'm back
What is the easiest explanation for SaaS? Just read this one
IDEA中如何连接数据库并显示数据库信息。
DNA 11. Identify mutation hotspots on the three-dimensional structure of tumor proteins (hotspot3d)
Dynamically debug JS code
Become a blogger in a year? To! Struggling oneself
DNA 6. Genome variation: complexheatmap
Game improvement of smart people: Chapter 3 Lesson 1: grid (grid position)
JS笔试题--浏览器内核
How to cultivate real data analysis thinking? Attached with practical cases
HDF5 library version mismatched error solution
Reptile analysis process from a rookie (code attached at the end of the text)
DNA 11. 识别肿瘤蛋白质三维结构上突变热点(HotSpot3D)
搞懂这些关键指标,数据分析起码少费一半力
一个极简美的登录框
c语言文件操作管理(上)
深度解析字符串和内存函数
通俗解释: IaaS,PaaS和SaaS的区别
Access数据库对象包括哪六个?Access与 Excel 最重要的区别是什么?
超全干货:数据可视化的设计总结,工具,技术要点统统都有